R
Part 2
It is very common these days to hear someone say “correlation does not mean causality.”
In essence, that is true.
Sometimes, there is causality even when we do not observe correlation.
The sailor is adjusting the rudder on a windy day to align the boat with the wind, but the boat is not changing direction. (Source: The Mixtape)
Note
In this example, the sailor is endogenously adjusting the course to balance the unobserved wind.

Imagine that you want to investigate the effect of Governance on Q
\(𝑸_{i} = α + 𝜷_{i} × Gov + Controls + error\)
All the issues in the next slides will make it not possible to infer that changing Gov will CAUSE a change in Q
That is, cannot infer causality

One source of bias is: reverse causation
Perhaps it is Q that causes Gov
OLS based methods do not tell the difference between these two betas:
\(𝑄_{i} = α + 𝜷_{i} × Gov + Controls + error\)
\(Gov_{i} = α + 𝜷_{i} × Q + Controls + error\)
If one Beta is significant, the other will most likely be significant too
You need a sound theory!
The second source of bias is: OVB
Imagine that you do not include an important “true” predictor of Q
Let’s say, long is: \(𝑸_{i} = 𝜶_{long} + 𝜷_{long}* gov_{i} + δ * omitted + error\)
But you estimate short: \(𝑸_{i} = 𝜶_{short} + 𝜷_{short}* gov_{i} + error\)
\(𝜷_{short}\) will be:
\(𝜷_{short} = 𝜷_{long}\) + bias
\(𝜷_{short} = 𝜷_{long}\) + relationship between omitted (omitted) and included (Gov) * effect of omitted in long (δ)
Thus, OVB is: \(𝜷_{short} – 𝜷_{long} = ϕ * δ\)
The third source of bias is: Specification error
Even if we could perfectly measure gov and all relevant covariates, we would not know for sure the functional form through which each influences q
Misspecification of x’s is similar to OVB
The fourth source of bias is: Signaling
Perhaps, some individuals are signaling the existence of an X without truly having it:
This is similar to the OVB because you cannot observe the full story
The fifth source of bias is: Simultaneity
Perhaps gov and some other variable x are determined simultaneously
Perhaps there is bidirectional causation, with q causing gov and gov also causing q
In both cases, OLS regression will provide a biased estimate of the effect
Also, the sign might be wrong
The sixth source of bias is: Heterogeneous effects
Maybe the causal effect of gov on q depends on observed and unobserved firm characteristics:
In such case, we may find a positive or negative relationship.
Neither is the true causal relationship
The seventh source of bias is: Construct validity
Some constructs (e.g. Corporate governance) are complex, and sometimes have conflicting mechanisms
We usually don’t know for sure what “good” governance is, for instance
It is common that we use imperfect proxies
They may poorly fit the underlying concept
The eighth source of bias is: Measurement error
“Classical” random measurement error for the outcome will inflate standard errors but will not lead to biased coefficients.
“Classical” random measurement error in x’s will bias coefficient estimates toward zero
The ninth source of bias is: Observation bias
This is analogous to the Hawthorne effect, in which observed subjects behave differently because they are observed
Firms which change gov may behave differently because their managers or employees think the change in gov matters, when in fact it has no direct effect
The tenth source of bias is: Interdependent effects
Imagine that a governance reform that will not affect share prices for a single firm might be effective if several firms adopt
Conversely, a reform that improves efficiency for a single firm might not improve profitability if adopted widely because the gains will be competed away
“One swallow doesn’t make a summer”
The eleventh source of bias is: Selection bias
If you run a regression with two types of companies
Without any matching method, these companies are likely not comparable
Thus, the estimated beta will contain selection bias
The bias can be either be positive or negative
It is similar to OVB
The twelfth source of bias is: Self-Selection
Self-selection is a type of selection bias
Usually, firms decide which level of governance they adopt
There are reasons why firms adopt high governance
It is like they “self-select” into the treatment.
Your coefficients will be biased.
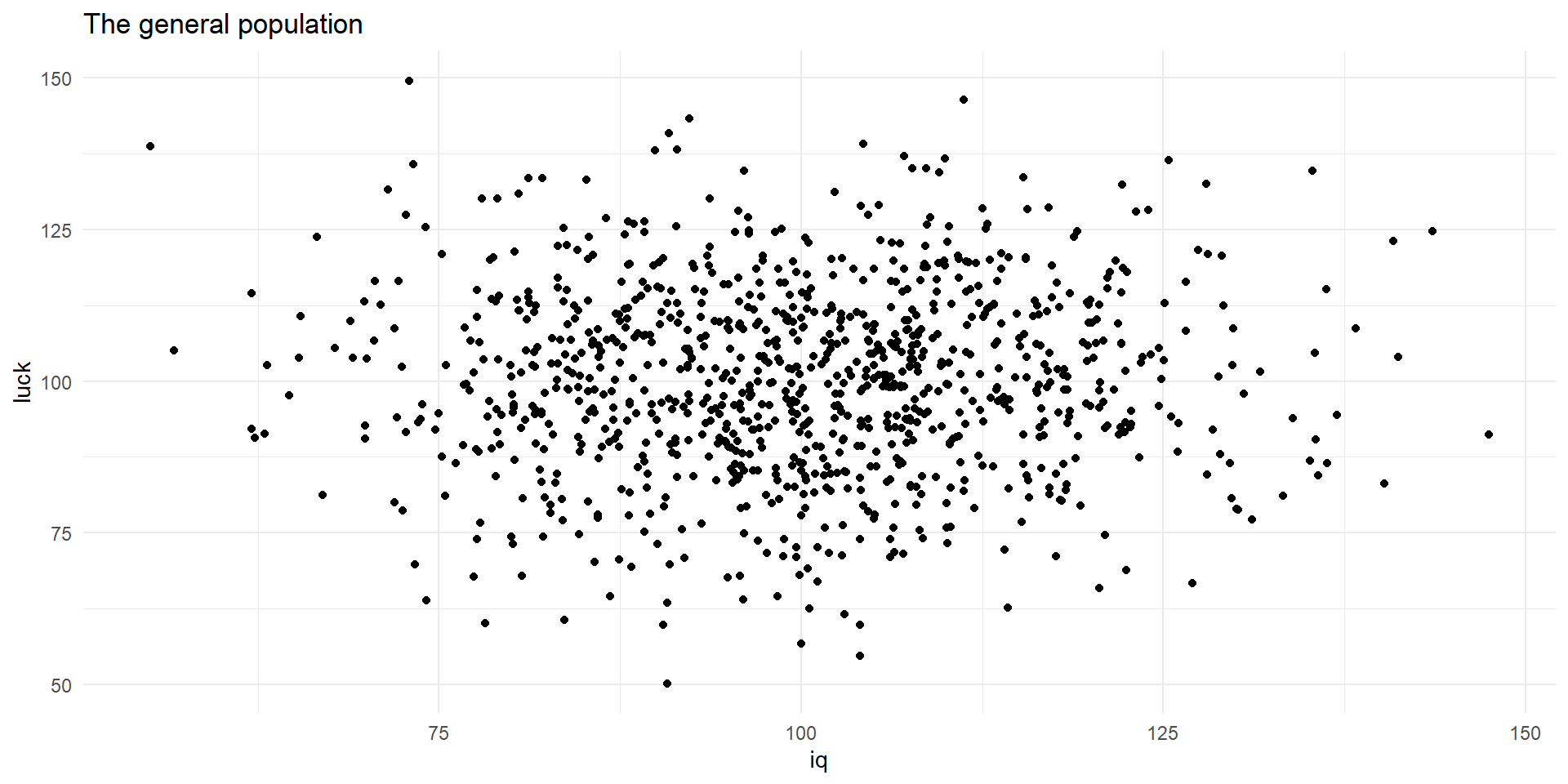
Let’s assume an arbitrary population.
Two variables describe the population: IQ and luck.
These variables are random and normally distributed.
Let’s say that after you reach a certain level of IQ and Luck, you become successful (i.e., upper-right quadrant).
Because you are interested in successful people, you only investigate such subsample.
A representation of the population
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
np.random.seed(100)
luck = np.random.normal(100, 15, 1000)
iq = np.random.normal(100, 15, 1000)
pop = pd.DataFrame({'luck': luck, 'iq': iq})
sns.set(style="whitegrid")
plt.figure(figsize=(8, 6))
plt.scatter(pop['iq'], pop['luck'])
plt.title("The general population")
plt.xlabel("IQ")
plt.ylabel("Luck")
plt.show()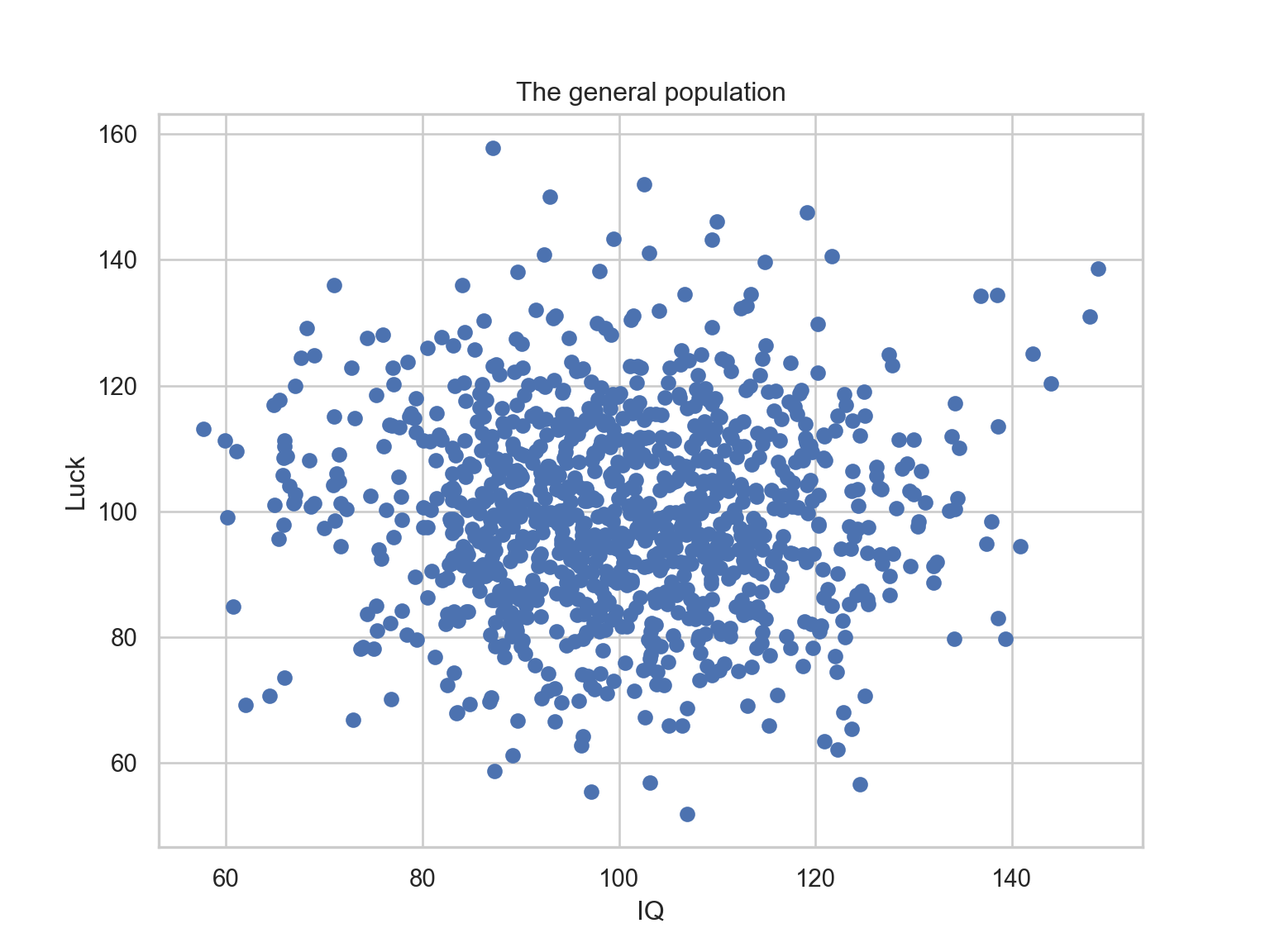
Important
Analyzing only successful people will suggest a negative correlation between luck and IQ.
library(data.table)
library(ggplot2)
set.seed(100)
luck <- rnorm(1000, 100, 15)
iq <- rnorm(1000, 100, 15)
pop <- data.frame(luck, iq)
pop$comb <- pop$luck + pop$iq
successfull <- pop[pop$comb > 240, ]
ggplot() +
geom_point(data = pop, aes(x = iq, y = luck)) +
geom_point(data = successfull, aes(x = iq, y = luck), color = "red") +
labs(title = "The general population & successful subpopulation") +
theme_minimal()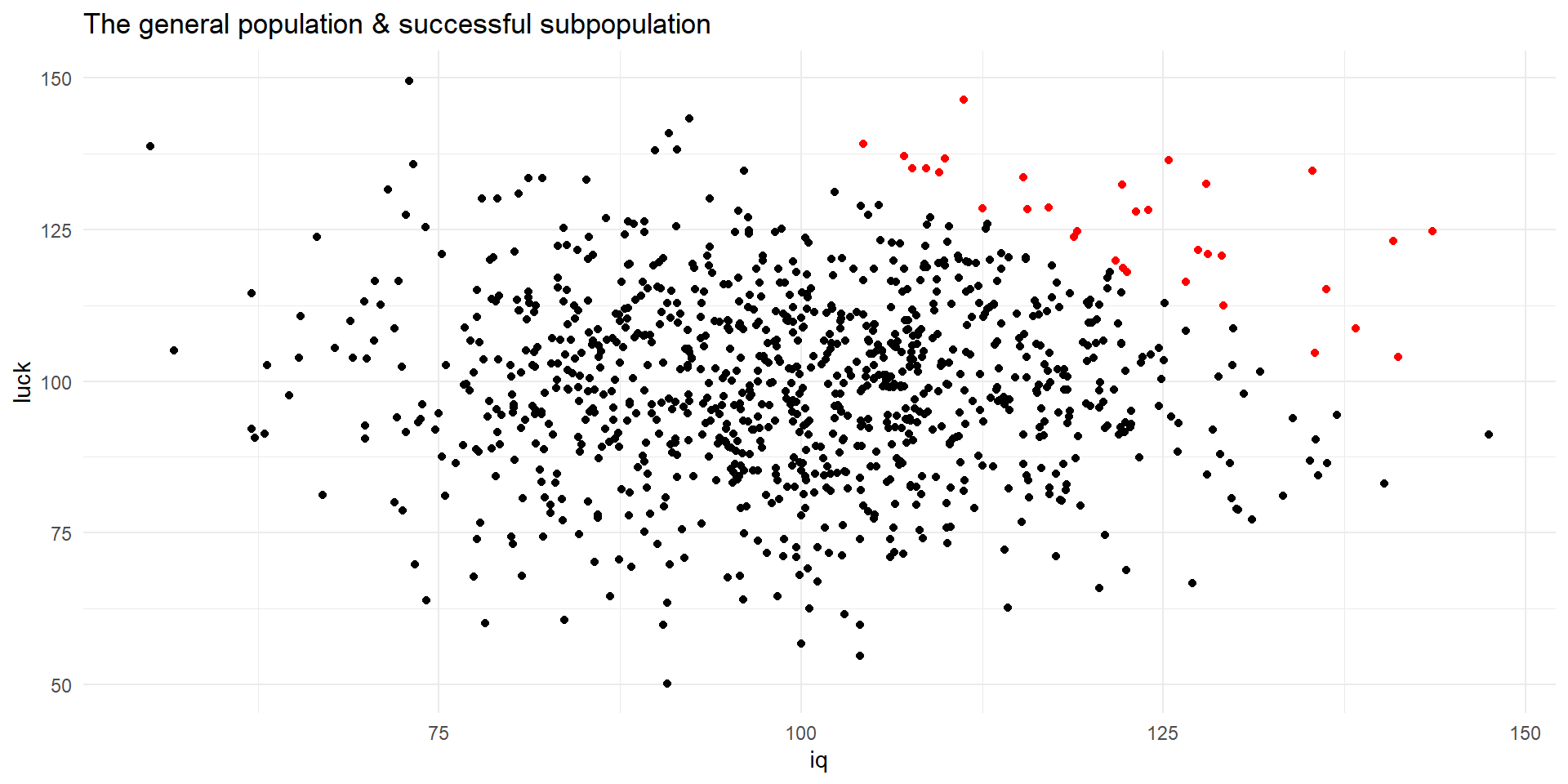
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
np.random.seed(100)
luck = np.random.normal(100, 15, 1000)
iq = np.random.normal(100, 15, 1000)
pop = pd.DataFrame({'luck': luck, 'iq': iq})
pop['comb'] = pop['luck'] + pop['iq']
successful = pop[pop['comb'] > 240]
sns.set(style="whitegrid") # Minimalistic theme similar to theme_minimal in ggplot2
plt.figure(figsize=(8, 6))
plt.scatter(pop['iq'], pop['luck'], label="General Population", alpha=0.5)
plt.scatter(successful['iq'], successful['luck'], color='red', label="Successful Subpopulation", alpha=0.7)
plt.title("The general population & successful subpopulation")
plt.xlabel("IQ")
plt.ylabel("Luck")
plt.legend()
plt.show()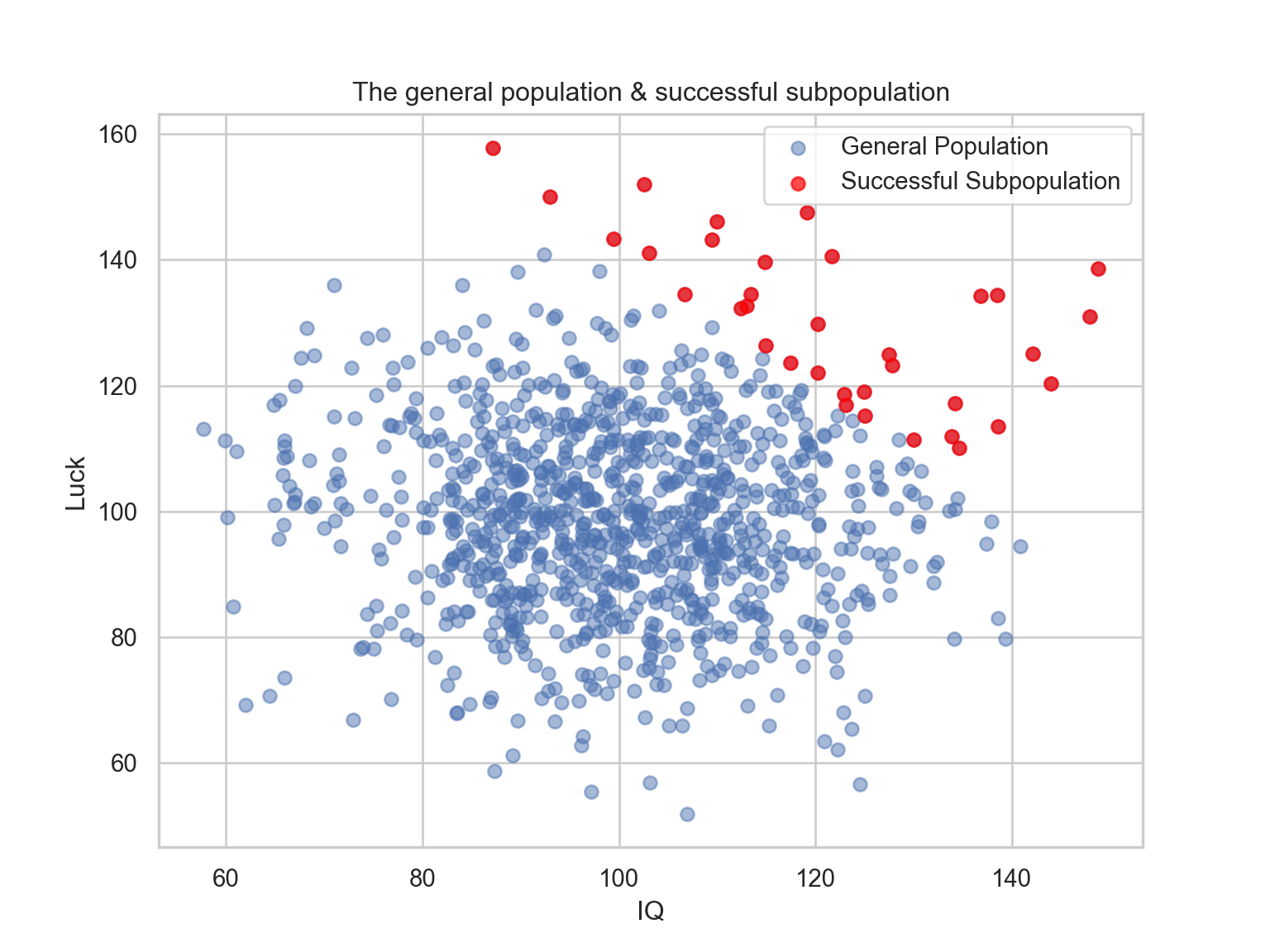
set seed 100
set obs 1000
gen luck = rnormal(100, 15)
gen iq = rnormal(100, 15)
gen comb = luck + iq
gen successful = comb > 240
twoway (scatter luck iq if successful == 0) (scatter luck iq if successful == 1, mcolor(red)) , title("The general population & successful subpopulation")
quietly graph export figs/collider2.svg, replaceNumber of observations (_N) was 0, now 1,000.Pesquisa quantitativa tem a parte quanti (métodos, modelos, etc.)…
… Mas talvez a parte mais importante seja o desenho da pesquisa (design empírico)!
P-Hacking
Artigo original aqui.
Publication bias
Artigo original aqui.
Crise de replicação
Artigo original aqui.

Henrique C. Martins [henrique.martins@fgv.br][Do not use without permission]